biolink-model
Schema and generated objects for biolink data model and upper ontology
This project is maintained by hsolbrig
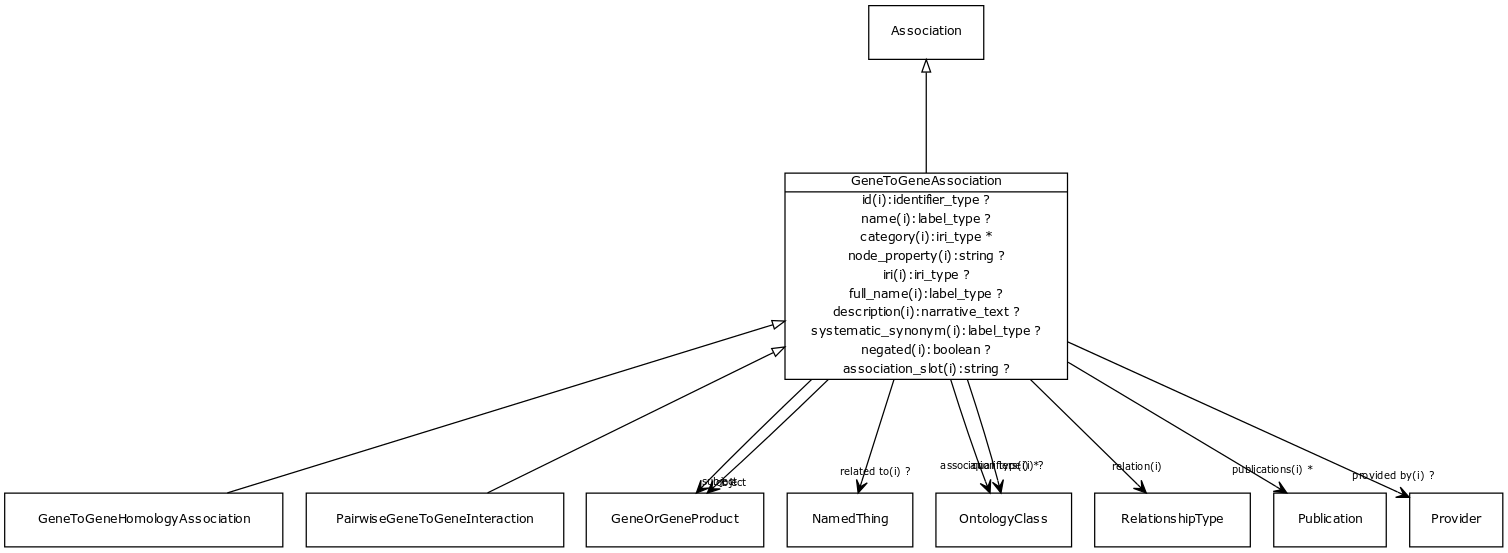
Class: gene to gene association
abstract parent class for different kinds of gene-gene or gene product to gene product relationships. Includes homology and interaction.
URI: http://w3id.org/biolink/vocab/GeneToGeneAssociation
Mappings
Inheritance
- is_a: Association - A typed association between two entities, supported by evidence
Children
- GeneToGeneHomologyAssociation - A homology association between two genes. May be orthology (in which case the species of subject and object should differ) or paralogy (in which case the species may be the same)
- PairwiseGeneToGeneInteraction - An interaction between two genes or two gene products. May be physical (e.g. protein binding) or genetic (between genes). May be symmetric (e.g. protein interaction) or directed (e.g. phosphorylation)
Used in
Fields
- gene to gene association.object
- Description: the object gene in the association. If the relation is symmetric, subject vs object is arbitrary. We allow a gene product to stand as proxy for the gene or vice versa
- range: GeneOrGeneProduct [required]
- Local
- gene to gene association.subject
- Description: the subject gene in the association. If the relation is symmetric, subject vs object is arbitrary. We allow a gene product to stand as proxy for the gene or vice versa
- range: GeneOrGeneProduct [required]
- Local
- association slot
- Description: any slot that relates an association to another entity
- range: string
- inherited from: Association
- association type
- Description: connects an association to the type of association (e.g. gene to phenotype)
- range: OntologyClass
- inherited from: Association
- category subsets: (translator_minimal)
- Description: Name of the high level ontology class in which this entity is categorized. Corresponds to the label for the biolink entity type class. In a neo4j database this MAY correspond to the neo4j label tag
- range: IriType*
- inherited from: NamedThing
- description subsets: (translator_minimal)
- Description: a human-readable description of a thing
- range: NarrativeText
- inherited from: NamedThing
- full name
- Description: a long-form human readable name for a thing
- range: LabelType
- inherited from: NamedThing
- id subsets: (translator_minimal)
- Description: A unique identifier for a thing. Must be either a CURIE shorthand for a URI or a complete URI
- range: IdentifierType
- inherited from: NamedThing
- iri subsets: (translator_minimal)
- Description: An IRI for the node. This is determined by the id using expansion rules.
- range: IriType
- inherited from: NamedThing
- name subsets: (translator_minimal)
- Description: A human-readable name for a thing
- range: LabelType
- inherited from: NamedThing
- negated
- Description: if set to true, then the association is negated i.e. is not true
- range: boolean
- inherited from: Association
- node property
- Description: A grouping for any property that holds between a node and a value
- range: string
- inherited from: NamedThing
- provided by
- Description: connects an association to the agent (person, organization or group) that provided it
- range: Provider
- inherited from: Association
- publications
- Description: connects an association to publications supporting the association
- range: Publication*
- inherited from: Association
- qualifiers
- Description: connects an association to qualifiers that modify or qualify the meaning of that association
- range: OntologyClass*
- inherited from: Association
- related to
- Description: A grouping for any relationship type that holds between any two things
- range: NamedThing
- inherited from: NamedThing
- relation
- Description: the relationship type by which a subject is connected to an object in an association
- range: RelationshipType [required]
- inherited from: Association
- systematic synonym
- Description: more commonly used for gene symbols in yeast
- range: LabelType
- inherited from: NamedThing